Abstract
Introduction: Myelodysplastic syndromes (MDS) are heterogeneous disorders caused by sequential accumulation of genetic lesions in haematopoietic stem cells (HSC). MDS are characterized by dysplasia, ineffective haematopoiesis and a propensity to evolve towards acute myeloid leukemia (AML). Based on the recent advances in next generation sequencing, our understanding of the genetic background of myeloid neoplasms has dramatically increased. However, clonal heterogeneity, genetic interactions, and clonal evolution remain enigmatic phenomena of oncogenesis and important investigational challenges to be addressed in the post-genomic era.
Aim and objectives: The aim of this project was to build a bioinformatic pipeline to integrate phosphoproteomic data with genetic information in order to characterize targettable kinase-activities of involved oncogenic pathways. Our objective was to establish a network analysis of the kinase-substrate relationship in myeloid cell lines first, before moving into primary cells. Here, we present data on the ongoing phosphoproteome characterization and the inferred kinase-activity from five myeloid cell lines.
Methods: The five myeloid cell lines K562 (erythroid), NB4 (promyelocytic), THP1 (monocytic), OCI-AML3 (myelomonocytic) and MOLM13 (monocytic) were used. These cell lines contain distinct oncogenes, such as BCR-ABL1, PML-RARalpha, MLL-MLLT3, mutated NPM1 and FLT3-ITD, respectively. Cytogenetic analysis and next generation sequencing, using a myeloid driver gene panel of Ion Torrent, were performed for genetic characterization (Table 1). Phosphoproteomes were enriched using Titanium-dioxide (TiO2) and samples were analyzed in triplicates by reversed-phase nano liquid chromatography coupled to tandem-mass spectrometry (nanoLC-MS2). Raw data was analyzed using MaxQuant software and further processed with R . For kinase-activity enrichment analysis (KAEA), we integrated all substrate-kinase datasets from five databases 1) PhosphoSitePlus, 2) Human Protein Reference Database, 3) Regulatory Network in Protein Phosphorylation, 4) The Signaling Network Open Resource and 5) phosphor.ELM . The SetRank R-package was employed for KAEA with p-value and FDR cutoffs set at 0.05.
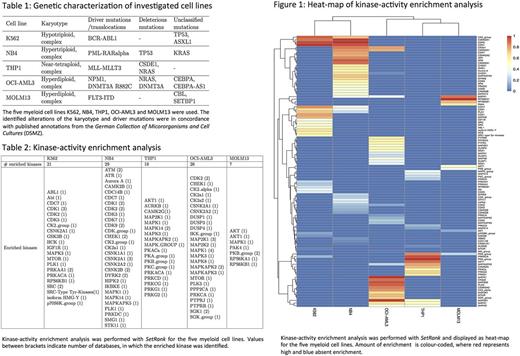
Results: We identified 15'698, 14'087, 13'969, 13'993 and 14'201 unique phosphopeptides corresponding to 3'536, 3'363, 3'411, 3'410 and 3'403 unique phosphoproteins, respectively.KAEA identified 77 different kinases across the five cell lines (Table 2). In the heat-map of the KAEA, phenotypically related cell lines clustered together and unique kinase-activity patterns emerged for each cell line (Figure 1). Two cell lines are driven by oncogenic kinases; K562 by BCR-ABL1 and MOLM13 by FLT3-ITD. An ABL1-kinase signal was detectable from 2 different databases in K562 as well as additional downstream kinases of ABL1, including mTOR and MAPK. We could not enrich for the FLT3-kinase in MOLM-13, due to lack of representation in the currently available substrate-kinase databases. However, our pipeline was able to identify the activity of downstream kinases of FLT3, including AKT1/PKB and MAPK1.
Conclusion: Our bioinformatic pipeline was able to detect unique phosphoproteins and enrich for kinase-activities in five distinct myeloid cell lines. Moreover, it reproduced an expected pattern of kinase-activities for two relevant driving oncogenic kinases. These promising results remain still limited by insufficient quantification and incompleteness of available databases. We expect to further improve on quantification and annotation by using heavy labeled cell lines (SILAC) as well as kinase-motif analysis, respectively. This may allow us to perform unsupervised characterization of involved oncogenic pathways and to proceed into the analysis of primary cells.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal